AppSAM ERC Starting Grant
AppSAM ERC Starting Grant
A Flexible Platform for the Application of SAM-dependent enzymes
AppSAM aims to develop an application platform for SAM-dependent enzymes that can be flexibly tailored to the application (and the enzyme) of choice. To achieve this broad applicability, different strategies, including in vitro and in vivo methodologies, as well as immobilisation techniques, were set up with a range of model enzymes.
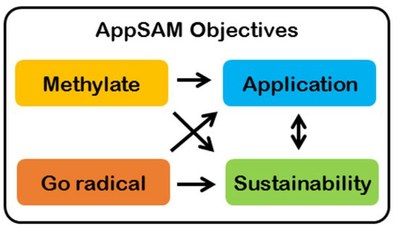
Connection between the establishment of enzymatic methylation and radical reactions and resulting sustainable applications.
S-Adenosylmethionine (SAM)-dependent enzymes play an important role in all kingdoms of life. SAM is used as a methyl donor for the methylation of small molecules, proteins, and ribonucleic acids by conventional methyltransferases. It is further the cofactor for radical SAM enzymes that can catalyse a multitude of complex reactions, including methylations at unactivated positions. In the wide field of epigenetics, methylations are also an important marker, an example are methylations of nucleobases that are introduced by conventional and radical SAM methyltransferases. The reactions catalysed by SAM-dependent enzymes are highly interesting for technical application, e.g. the synthesis of selectively methylated building blocks for pharmaceuticals. Often, the corresponding synthetically-chemical version of the reaction uses highly toxic and cancerogenic compounds and is rather unselective. In the AppSAM project, we developed strategies to integrate SAM-dependent enzymes in multi-enzyme cascades to facilitate their handling and efficiency for chemical synthesis. These systems are also useful for the mechanistic-functional investigation of SAM-dependent enzymes, e.g. from epigenetic pathways or natural product biosynthesis, using modified (co-)substrates that can be produced in situ. In addition to in vitro multienzyme systems, different in vivo systems have been explored with a range of enzyme/ substrate combinations.
A milestone was the successful implementation of radical SAM regeneration, applicable to all SAM-dependent enzymes. The flexibility of such regeneration systems was illustrated by the transfer of an ethyl group with a cobalamin-dependent radical SAM methyltransferase using S-adenosylethionine as a cofactor
(doi 10.1002/cbic.202300133, doi 10.1002/ange.202204198).
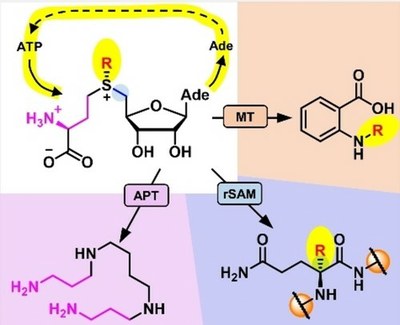
Biomimetic S-Adenosylmethionine Regeneration Starting from Multiple Byproducts Enables Biocatalytic Alkylation with Radical SAM Enzymes (Gericke et al., 2023).
Overexpression of a single gene encoding a methyltransferase (MT), including in vivo biotransformation and purification of the MT product, was established. This approach was then compared with a classical in vitro upscaling method (doi 10.1002/cctc.202300930).
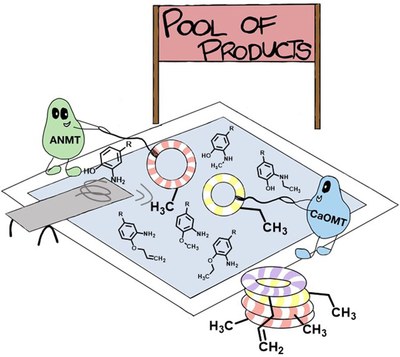
Expanding the Substrate Scope of N- and O-Methyltransferases from Plants for Chemoselective Alkylation
(Jockmann et al., 2023).
Our research group received the RSC "Outstanding Paper Award 2021" for a bicyclic SAM regeneration system applicable with different nucleotides and nucleosides as cofactor building blocks.
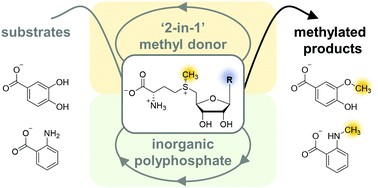
A bicyclic S-adenosylmethionine regeneration system applicable with different nucleosides or nucleotides as cofactor building blocks (
Publications
Peer Reviewed Articles
DOI: 10.1038/s42003-024-06078-9.
DOI: 10.1002/cctc.202300930.
M. K. F. Mohr, R. Saleem-Batcha, N. V. Cornelissen, J. N. Andexer, Enzymatic Synthesis of L-Methionine Analogues and Application in a Methyltransferase Catalysed Alkylation Cascade, Chemistry – A European Journal 2023, DOI: 10.1002/chem.20230150
L. Gericke, D. Mhaindarkar, L. C. Karst, S. Jahn, M. Kuge, M. K. F. Mohr, J. Gagsteiger, N. V. Cornelissen, X. Wen, S. Mordhorst, H. J. Jessen, A. Rentmeister, F. P. Seebeck, G. Layer, C. Loenarz, J. N. Andexer, Biomimetic S-Adenosylmethionine Regeneration Starting from Multiple Byproducts Enables Biocatalytic Alkylation with Radical SAM Enzymes, ChemBioChem 2023, DOI: 10.1002/cbic.202300133.
J. Gagsteiger, S. Jahn, L. Heidinger, L. Gericke, J. N. Andexer, T. Friedrich, C. Loenarz, G. Layer, A Cobalamin-Dependent Radical SAM Enzyme Catalyzes the Unique Cα-Methylation of Glutamine in Methyl-Coenzyme M Reductase, Angew. Chem. Int. Ed. 2022, 134, e202204198, DOI: 10.1002/ange.202204198.
D. Popadić, D. Mhaindarkar, M. H. N. Dang Thai, H. C. Hailes, S. Mordhorst, J. N. Andexer, A bicyclic S-adenosylmethionine regeneration system applicable with different nucleosides or nucleotides as cofactor building blocks. RSC Chem. Biol. 2021, DOI: 10.1039/D1CB00033K.
ERC Team Members
Dr. Raspudin Saleem-Batcha
Dr. Philipp Germer
Dr. Lukas Karst
Dr. Dipali Mhaindarkar
Dr. Silja Mordhorst
Lukas Gericke
Lars-Hendrik Köppl
Michael Mohr
Mike Dhang Thai

